This article possibly contains original research. (July 2015) |
| Ankyrin repeat domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | Ank | ||||||||||
| Pfam | PF00023 | ||||||||||
| InterPro | IPR002110 | ||||||||||
| SMART | SM00248 | ||||||||||
| PROSITE | PDOC50088 | ||||||||||
| SCOP2 | 1awc / SCOPe / SUPFAM | ||||||||||
| |||||||||||
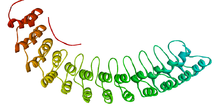
The ankyrin repeat is a 33-residue motif in proteins consisting of two alpha helices separated by loops, first discovered in signaling proteins in yeast Cdc10 and Drosophila Notch. Domains consisting of ankyrin tandem repeats mediate protein–protein interactions and are among the most common structural motifs in known proteins. They appear in bacterial, archaeal, and eukaryotic proteins, but are far more common in eukaryotes. Ankyrin repeat proteins, though absent in most viruses, are common among poxviruses. Most proteins that contain the motif have four to six repeats, although its namesake ankyrin contains 24, and the largest known number of repeats is 34, predicted in a protein expressed by Giardia lamblia.[2]
Ankyrin repeats typically fold together to form a single, linear solenoid structure called ankyrin repeat domains. These domains are one of the most common protein–protein interaction platforms in nature. They occur in a large number of functionally diverse proteins, mainly from eukaryotes. The few known examples from prokaryotes and viruses may be the result of horizontal gene transfers.[3] The repeat has been found in proteins of diverse function such as transcriptional initiators, cell cycle regulators, cytoskeletal, ion transporters, and signal transducers. The ankyrin fold appears to be defined by its structure rather than its function, since there is no specific sequence or structure that is universally recognised by it.
Considering the atomic structures of individual ankyrin repeats, the loop is often a type 1 beta bulge loop, while both alpha-helices commonly have a Schellman loop at their N-terminus.
- ^ PDB: 1N11; Michaely P, Tomchick DR, Machius M, Anderson RG (December 2002). "Crystal structure of a 12 ANK repeat stack from human ANK1". EMBO J. 21 (23): 6387–96. doi:10.1093/emboj/cdf651. PMC 136955. PMID 12456646.
- ^ Mosavi L, Cammett T, Desrosiers D, Peng Z (2004). "The ankyrin repeat as molecular architecture for protein recognition". Protein Sci. 13 (6): 1435–48. doi:10.1110/ps.03554604. PMC 2279977. PMID 15152081. Archived from the original on 2004-09-07.
- ^ Bork P (December 1993). "Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally?". Proteins. 17 (4): 363–74. doi:10.1002/prot.340170405. PMID 8108379. S2CID 35224626.