| CBS domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
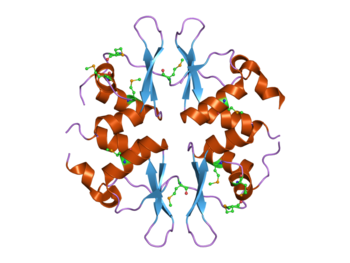 Structure of the yeast SNF4 protein that contains four CBS domains.[1] This protein is part of the AMP-activated protein kinase (AMPK) complex. | |||||||||||
| Identifiers | |||||||||||
| Symbol | CBS | ||||||||||
| Pfam | PF00571 | ||||||||||
| InterPro | IPR000644 | ||||||||||
| SMART | CBS | ||||||||||
| PROSITE | PS51371 | ||||||||||
| SCOP2 | 1zfj / SCOPe / SUPFAM | ||||||||||
| CDD | cd02205 | ||||||||||
| |||||||||||
In molecular biology, the CBS domain is a protein domain found in a range of proteins in all species from bacteria to humans. It was first identified as a conserved sequence region in 1997 and named after cystathionine beta synthase, one of the proteins it is found in.[2] CBS domains are also found in a wide variety of other proteins such as inosine monophosphate dehydrogenase,[3] voltage gated chloride channels[4][5][6][7][8] and AMP-activated protein kinase (AMPK).[9][10] CBS domains regulate the activity of associated enzymatic and transporter domains in response to binding molecules with adenosyl groups such as AMP and ATP, or s-adenosylmethionine.[11]
- ^ PDB: 2nye; Rudolph MJ, Amodeo GA, Iram SH, Hong SP, Pirino G, Carlson M, Tong L (January 2007). "Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding". Structure. 15 (1): 65–74. doi:10.1016/j.str.2006.11.014. PMID 17223533.
- ^ Bateman A (January 1997). "The structure of a domain common to archaebacteria and the homocystinuria disease protein". Trends Biochem. Sci. 22 (1): 12–3. doi:10.1016/S0968-0004(96)30046-7. PMID 9020585.
- ^ Ignoul S, Eggermont J (December 2005). "CBS domains: structure, function, and pathology in human proteins". Am. J. Physiol., Cell Physiol. 289 (6): C1369–78. doi:10.1152/ajpcell.00282.2005. PMID 16275737.
- ^ Ponting CP (March 1997). "CBS domains in CIC chloride channels implicated in myotonia and nephrolithiasis (kidney stones)". J. Mol. Med. 75 (3): 160–3. PMID 9106071.
- ^ Meyer S, Dutzler R (February 2006). "Crystal structure of the cytoplasmic domain of the chloride channel ClC-0". Structure. 14 (2): 299–307. doi:10.1016/j.str.2005.10.008. PMID 16472749.
- ^ Yusef YR, Zúñiga L, Catalán M, Niemeyer MI, Cid LP, Sepúlveda FV (April 2006). "Removal of gating in voltage-dependent ClC-2 chloride channel by point mutations affecting the pore and C-terminus CBS-2 domain". J. Physiol. 572 (Pt 1): 173–81. doi:10.1113/jphysiol.2005.102392. PMC 1779660. PMID 16469788.
- ^ Markovic S, Dutzler R (June 2007). "The structure of the cytoplasmic domain of the chloride channel ClC-Ka reveals a conserved interaction interface". Structure. 15 (6): 715–25. doi:10.1016/j.str.2007.04.013. PMID 17562318.
- ^ Meyer S, Savaresi S, Forster IC, Dutzler R (January 2007). "Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5". Nat. Struct. Mol. Biol. 14 (1): 60–7. doi:10.1038/nsmb1188. PMID 17195847. S2CID 20733119.
- ^ Day P, Sharff A, Parra L, et al. (May 2007). "Structure of a CBS-domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP and ZMP". Acta Crystallogr. D. 63 (Pt 5): 587–96. doi:10.1107/S0907444907009110. PMID 17452784.
- ^ Rudolph MJ, Amodeo GA, Iram SH, et al. (January 2007). "Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding". Structure. 15 (1): 65–74. doi:10.1016/j.str.2006.11.014. PMID 17223533.
- ^ Kemp BE (January 2004). "Bateman domains and adenosine derivatives form a binding contract". J. Clin. Invest. 113 (2): 182–4. doi:10.1172/JCI20846. PMC 311445. PMID 14722609.