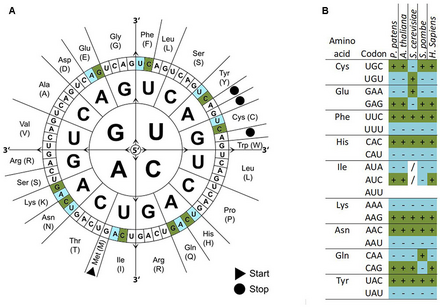
Codon usage bias refers to differences in the frequency of occurrence of synonymous codons in coding DNA. A codon is a series of three nucleotides (a triplet) that encodes a specific amino acid residue in a polypeptide chain or for the termination of translation (stop codons).
There are 64 different codons (61 codons encoding for amino acids and 3 stop codons) but only 20 different translated amino acids. The overabundance in the number of codons allows many amino acids to be encoded by more than one codon. Because of such redundancy it is said that the genetic code is degenerate. The genetic codes of different organisms are often biased towards using one of the several codons that encode the same amino acid over the others—that is, a greater frequency of one will be found than expected by chance. How such biases arise is a much debated area of molecular evolution. Codon usage tables detailing genomic codon usage bias for organisms in GenBank and RefSeq can be found in the HIVE-Codon Usage Tables (HIVE-CUTs) project[dead link],[1] which contains two distinct databases, CoCoPUTs and TissueCoCoPUTs. Together, these two databases provide comprehensive, up-to-date codon, codon pair and dinucleotide usage statistics for all organisms with available sequence information and 52 human tissues, respectively.[2][3]
It is generally acknowledged that codon biases reflect the contributions of 3 main factors: GC-biased gene conversion that favors GC-ending codons in diploid organisms, arrival biases reflecting mutational preferences (typically favoring AT-ending codons), and natural selection for codons that are favorable in regard to translation.[4] [5] [6] Optimal codons in fast-growing microorganisms, like Escherichia coli or Saccharomyces cerevisiae (baker's yeast), reflect the composition of their respective genomic transfer RNA (tRNA) pool.[7] It is thought that optimal codons help to achieve faster translation rates and high accuracy. As a result of these factors, translational selection is expected to be stronger in highly expressed genes, as is indeed the case for the above-mentioned organisms.[8][9] In other organisms that do not show high growing rates or that present small genomes, codon usage optimization is normally absent, and codon preferences are determined by the characteristic mutational biases seen in that particular genome. Examples of this are Homo sapiens (human) and Helicobacter pylori.[10][11] Organisms that show an intermediate level of codon usage optimization include Drosophila melanogaster (fruit fly), Caenorhabditis elegans (nematode worm), Strongylocentrotus purpuratus (sea urchin), and Arabidopsis thaliana (thale cress).[12] Several viral families (herpesvirus, lentivirus, papillomavirus, polyomavirus, adenovirus, and parvovirus) are known to encode structural proteins that display heavily skewed codon usage compared to the host cell. The suggestion has been made that these codon biases play a role in the temporal regulation of their late proteins.[13]
The nature of the codon usage-tRNA optimization has been fiercely debated. It is not clear whether codon usage drives tRNA evolution or vice versa. At least one mathematical model has been developed where both codon usage and tRNA expression co-evolve in feedback fashion (i.e., codons already present in high frequencies drive up the expression of their corresponding tRNAs, and tRNAs normally expressed at high levels drive up the frequency of their corresponding codons). However, this model does not seem to yet have experimental confirmation. Another problem is that the evolution of tRNA genes has been a very inactive area of research.[citation needed]
- ^ Athey, John; Alexaki, Aikaterini; Osipova, Ekaterina; Rostovtsev, Alexandre; Santana-Quintero, Luis V.; Katneni, Upendra; Simonyan, Vahan; Kimchi-Sarfaty, Chava (2017-09-02). "A new and updated resource for codon usage tables". BMC Bioinformatics. 18 (391): 391. doi:10.1186/s12859-017-1793-7. PMC 5581930. PMID 28865429.
- ^ Alexaki, Aikaterini; Kames, Jacob; Holcomb, David D.; Athey, John; Santana-Quintero, Luis V.; Lam, Phuc Vihn Nguyen; Hamasaki-Katagiri, Nobuko; Osipova, Ekaterina; Simonyan, Vahan; Bar, Haim; Komar, Anton A.; Kimchi-Sarfaty, Chava (June 2019). "Codon and Codon-Pair Usage Tables (CoCoPUTs): Facilitating Genetic Variation Analyses and Recombinant Gene Design". Journal of Molecular Biology. 431 (13): 2434–2441. doi:10.1016/j.jmb.2019.04.021. PMID 31029701. S2CID 139104807.
- ^ Kames, Jacob; Alexaki, Aikaterini; Holcomb, David D.; Santana-Quintero, Luis V.; Athey, John C.; Hamasaki-Katagiri, Nobuko; Katneni, Upendra; Golikov, Anton; Ibla, Juan C.; Bar, Haim; Kimchi-Sarfaty, Chava (January 2020). "TissueCoCoPUTs: Novel Human Tissue-Specific Codon and Codon-Pair Usage Tables Based on Differential Tissue Gene Expression". Journal of Molecular Biology. 432 (11): 3369–3378. doi:10.1016/j.jmb.2020.01.011. PMID 31982380.
- ^ P. Shah and M. A. Gilchrist (2011). "Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift". Proceedings of the National Academy of Sciences of the United States of America. 108 (25): 10231–6. doi:10.1073/pnas.1016719108. PMC 3121864. PMID 21646514.
- ^ L. Duret and N. Galtier (2009). "Biased gene conversion and the evolution of mammalian genomic landscapes". Annu Rev Genomics Hum Genet. 10: 285–311. doi:10.1146/annurev-genom-082908-150001.
- ^ N. Galtier, C. Roux, M. Rousselle, J. Romiguier, E. Figuet, S. Glemin, N. Bierne and L. Duret (2018). "Codon Usage Bias in Animals: Disentangling the Effects of Natural Selection, Effective Population Size, and GC-Biased Gene Conversion". Mol Biol Evol. 35 (5): 1092–1103. doi:10.1093/molbev/msy015. hdl:20.500.12210/34500.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Dong, Hengjiang; Nilsson, Lars; Kurland, Charles G. (1996). "Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates". Journal of Molecular Biology. 260 (5): 649–663. doi:10.1006/jmbi.1996.0428. ISSN 0022-2836. PMID 8709146.
- ^ Sharp, Paul M.; Stenico, Michele; Peden, John F.; Lloyd, Andrew T. (1993). "Codon usage: mutational bias, translational selection, or both?". Biochem. Soc. Trans. 21 (4): 835–841. doi:10.1042/bst0210835. PMID 8132077. S2CID 8582630.
- ^ Kanaya, Shigehiko; Yamada, Yuko; Kudo, Yoshihiro; Ikemura, Toshimichi (1999). "Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis". Gene. 238 (1): 143–155. doi:10.1016/s0378-1119(99)00225-5. ISSN 0378-1119. PMID 10570992.
- ^ Atherton, John C.; Sharp, Paul M.; Lafay, Bénédicte (2000-04-01). "Absence of translationally selected synonymous codon usage bias in Helicobacter pylori". Microbiology. 146 (4): 851–860. doi:10.1099/00221287-146-4-851. ISSN 1350-0872. PMID 10784043.
- ^ Bornelöv, Susanne; Selmi, Tommaso; Flad, Sophia; Dietmann, Sabine; Frye, Michaela (2019-06-07). "Codon usage optimization in pluripotent embryonic stem cells". Genome Biology. 20 (1): 119. doi:10.1186/s13059-019-1726-z. ISSN 1474-760X. PMC 6555954. PMID 31174582.
- ^ Duret, Laurent (2000). "tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes". Trends in Genetics. 16 (7): 287–289. doi:10.1016/s0168-9525(00)02041-2. ISSN 0168-9525. PMID 10858656.
- ^ Shin, Young C.; Bischof, Georg F.; Lauer, William A.; Desrosiers, Ronald C. (2015-09-10). "Importance of codon usage for the temporal regulation of viral gene expression". Proceedings of the National Academy of Sciences. 112 (45): 14030–14035. Bibcode:2015PNAS..11214030S. doi:10.1073/pnas.1515387112. PMC 4653223. PMID 26504241.