 | |
| Content | |
|---|---|
| Description | Single Nucleotide Polymorphism Database |
| Organisms | Homo sapiens |
| Contact | |
| Research center | National Center for Biotechnology Information |
| Primary citation | PMID 21097890 |
| Release date | 1998 |
| Access | |
| Data format | ASN.1, Fasta, XML |
| Website | ncbi |
| Download URL | ftp://ftp.ncbi.nih.gov/snp/ |
| Web service URL | EUtils SOAP |
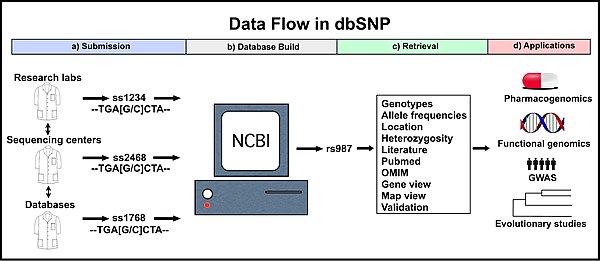
The Single Nucleotide Polymorphism Database[1] (dbSNP) is a free public archive for genetic variation within and across different species developed and hosted by the National Center for Biotechnology Information (NCBI) in collaboration with the National Human Genome Research Institute (NHGRI). Although the name of the database implies a collection of one class of polymorphisms only (i.e., single nucleotide polymorphisms (SNPs)), it in fact contains a range of molecular variation: (1) SNPs, (2) short deletion and insertion polymorphisms (indels/DIPs), (3) microsatellite markers or short tandem repeats (STRs), (4) multinucleotide polymorphisms (MNPs), (5) heterozygous sequences, and (6) named variants.[2] The dbSNP accepts apparently neutral polymorphisms, polymorphisms corresponding to known phenotypes, and regions of no variation. It was created in September 1998 to supplement GenBank, NCBI’s collection of publicly available nucleic acid and protein sequences.[2]
In 2017, NCBI stopped support for all non-human organisms in dbSNP.[3] As of build 153 (released in August 2019), dbSNP had amassed nearly 2 billion submissions representing more than 675 million distinct variants for Homo sapiens.
- ^ Wheeler DL, Barrett T, Benson DA, et al. (January 2007). "Database resources of the National Center for Biotechnology Information". Nucleic Acids Res. 35 (Database issue): D5–12. doi:10.1093/nar/gkl1031. PMC 1781113. PMID 17170002.
- ^ a b Sherry ST, Ward M; Sirotkin, K. (1999). "dbSNP - database for single nucleotide polymorphisms and other classes of minor genetic variation". Genome Research. 9 (8): 677–679. doi:10.1101/gr.9.8.677. PMID 10447503. S2CID 10775908.
- ^ "Phasing out support for non-human genome organism data in dbSNP and dbVar". 2017-05-09. Retrieved 9 July 2017.