| Nuclease S1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.1.30.1 | ||||||||
| CAS no. | 37288-25-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
| S1-P1 nuclease | |||||||||
|---|---|---|---|---|---|---|---|---|---|
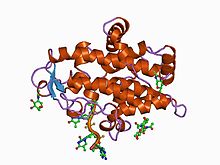 P1 nuclease in complex with a substrate analog | |||||||||
| Identifiers | |||||||||
| Symbol | S1-P1_nuclease | ||||||||
| Pfam | PF02265 | ||||||||
| Pfam clan | CL0368 | ||||||||
| InterPro | IPR003154 | ||||||||
| SCOP2 | 1ak0 / SCOPe / SUPFAM | ||||||||
| CDD | cd11010 | ||||||||
| |||||||||
Nuclease S1 (EC 3.1.30.1) is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalyses the following chemical reaction
- Endonucleolytic cleavage to 5'-phosphomononucleotide and 5'-phosphooligonucleotide end-products
Although its primary substrate is single-stranded, it can also occasionally introduce single-stranded breaks in double-stranded DNA or RNA, or DNA-RNA hybrids. The enzyme hydrolyses single stranded region in duplex DNA such as loops or gaps. It also cleaves a strand opposite a nick on the complementary strand. It has no sequence specificity.
Well-known versions include S1 found in Aspergillus oryzae (yellow koji mold) and Nuclease P1 found in Penicillium citrinum. Members of the S1/P1 family are found in both prokaryotes and eukaryotes and are thought to be associated in programmed cell death and also in tissue differentiation. Furthermore, they are secreted extracellular, that is, outside of the cell. Their function and distinguishing features mean they have potential in being exploited in the field of biotechnology.