| |
| Names | |
|---|---|
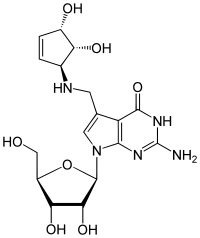
| IUPAC name
7-({[(1S,4S,5R)-4,5-Dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-carbaguanosine
| |
| Systematic IUPAC name
2-Amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one | |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
PubChem CID
|
|
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C17H23N5O7 | |
| Molar mass | 409.399 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Queuosine is a modified nucleoside that is present in certain tRNAs in bacteria and eukaryotes.[1][2] It contains the nucleobase queuine. Originally identified in E. coli, queuosine was found to occupy the first anticodon position of tRNAs for histidine, aspartic acid, asparagine and tyrosine.[3] The first anticodon position pairs with the third "wobble" position in codons, and queuosine improves accuracy of translation compared to guanosine.[4][5][6] Synthesis of queuosine begins with GTP. In bacteria, three structurally unrelated classes of riboswitch are known to regulate genes that are involved in the synthesis or transport of pre-queuosine1, a precursor to queuosine: PreQ1-I riboswitches, PreQ1-II riboswitches and PreQ1-III riboswitches.
Queuosine biosynthesis genes have also been found on phage genomes and may be involved in protection from genome degradation by the host.[7]
- ^ Iwata-Reuyl D (2003). "Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA". Bioorg. Chem. 31 (1): 24–43. doi:10.1016/S0045-2068(02)00513-8. PMID 12697167.
- ^ Morris RC, Elliott MS (2001). "Queuosine modification of tRNA: a case for convergent evolution". Mol. Genet. Metab. 74 (1–2): 147–159. doi:10.1006/mgme.2001.3216. PMID 11592812.
- ^ Harada F, Nishimura S (1972). "Possible anticodon sequences of tRNAHis, tRNAAsm, and tRNAAsp from Escherichia coli B. Universal presence of nucleoside Q in the first position of the anticondons of these transfer ribonucleic acids". Biochemistry. 11 (2): 301–308. doi:10.1021/bi00752a024. PMID 4550561.
- ^ Bienz M, Kubli E (1981). "Wild-type tRNATyrG reads the TMV RNA stop codon, but Q base-modified tRNATyrQ does not". Nature. 294 (5837): 188–190. Bibcode:1981Natur.294..188B. doi:10.1038/294188a0. PMID 29451243. S2CID 204999725.
- ^ Meier F, Suter B, Grosjean H, Keith G, Kubli E (1985). "Queuosine modification of the wobble base in tRNAHis influences 'in vivo' decoding properties". EMBO J. 4 (3): 823–827. doi:10.1002/j.1460-2075.1985.tb03704.x. PMC 554263. PMID 2988936.
- ^ Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Björk GR (2001). "Improvement of reading frame maintenance is a common function for several tRNA modifications". EMBO J. 20 (17): 4863–4873. doi:10.1093/emboj/20.17.4863. PMC 125605. PMID 11532950.
- ^ "Comparative genomics of bacteriophage of the genus Seuratvirus". Genome Biol Evol. 10: 72–76. 2022-04-07. doi:10.1063/5.0085058.7. Retrieved 2022-08-18.