| ATP Synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
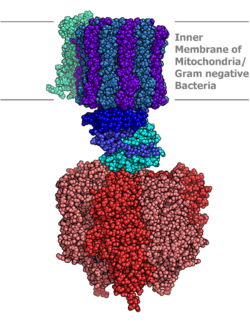 Molecular model of ATP synthase determined by X-ray crystallography. Stator is not shown here. | |||||||||
| Identifiers | |||||||||
| EC no. | 7.1.2.2 | ||||||||
| CAS no. | 9000-83-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
ATP synthase is an enzyme that catalyzes the formation of the energy storage molecule adenosine triphosphate (ATP) using adenosine diphosphate (ADP) and inorganic phosphate (Pi). ATP synthase is a molecular machine. The overall reaction catalyzed by ATP synthase is:
- ADP + Pi + 2H+out ⇌ ATP + H2O + 2H+in
ATP synthase lies across a cellular membrane and forms an aperture that protons can cross from areas of high concentration to areas of low concentration, imparting energy for the synthesis of ATP. This electrochemical gradient is generated by the electron transport chain and allows cells to store energy in ATP for later use. In prokaryotic cells ATP synthase lies across the plasma membrane, while in eukaryotic cells it lies across the inner mitochondrial membrane. Organisms capable of photosynthesis also have ATP synthase across the thylakoid membrane, which in plants is located in the chloroplast and in cyanobacteria is located in the cytoplasm.
Eukaryotic ATP synthases are F-ATPases, running "in reverse" for an ATPase. This article deals mainly with this type. An F-ATPase consists of two main subunits, FO and F1, which has a rotational motor mechanism allowing for ATP production.[1][2]
- ^ Okuno D, Iino R, Noji H (June 2011). "Rotation and structure of FoF1-ATP synthase". Journal of Biochemistry. 149 (6): 655–664. doi:10.1093/jb/mvr049. PMID 21524994.
- ^ Junge W, Nelson N (June 2015). "ATP synthase". Annual Review of Biochemistry. 84: 631–657. doi:10.1146/annurev-biochem-060614-034124. PMID 25839341.