| Chlamydomonas | |
|---|---|
| |
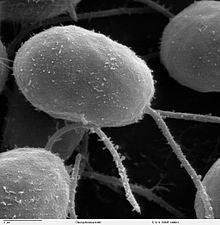
| SEM image of flagellated Chlamydomonas - Genus of Algae (10,000×) | |
| Scientific classification | |
| Clade: | Viridiplantae |
| Division: | Chlorophyta |
| Class: | Chlorophyceae |
| Order: | Chlamydomonadales |
| Family: | Chlamydomonadaceae |
| Genus: | Chlamydomonas Ehrenb. |
| Species | |
|
See text | |



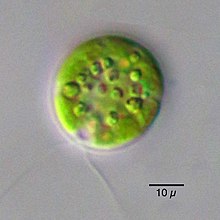
Chlamydomonas (/ˌklæmɪˈdɒmənəs, -dəˈmoʊ-/ KLAM-ih-DOM-ə-nəs, -də-MOH-) is a genus of green algae consisting of about 150 species[2] of unicellular flagellates, found in stagnant water and on damp soil, in freshwater, seawater, and even in snow as "snow algae".[3] Chlamydomonas is used as a model organism for molecular biology, especially studies of flagellar motility and chloroplast dynamics, biogenesis, and genetics. One of the many striking features of Chlamydomonas is that it contains ion channels (channelrhodopsins) that are directly activated by light. Some regulatory systems of Chlamydomonas are more complex than their homologs in Gymnosperms, with evolutionarily related regulatory proteins being larger and containing additional domains.[4]
Molecular phylogeny studies indicated that the traditional genus Chlamydomonas as defined using morphological data, was polyphyletic within Volvocales. Many species were subsequently reclassified (e.g., Oogamochlamys, Lobochlamys), and many other "Chlamydomonas" s.l. lineages are still to be reclassified.[5][6][7]
- ^ Hazen, Tracy E. 1922. The phylogeny of the genus Brachiomonas. Bulletin of the Torrey Botanical Club. 49(4):75-92, with two plates.
- ^ Smith, G. M. 1955 Cryptogamic Botany Volume 1. Algae and Fungi McGraw-Hill Book Company Inc
- ^ Hoham, R. W., Bonome, T. A., Martin, C. W. and Leebens-mack, J. H. 2002. A combined 18S rDNA and rbcL phylogenetic analysis of Chloromonas and Chlamydomonas (Chlorophyceae, Volvocales ) emphasizing snow and other cold-temperature habitats. Journal of Phycology, 38: 1051–1064. [1]
- ^ A Falciatore, L Merendino, F Barneche, M Ceol, R Meskauskiene, K Apel, JD Rochaix (2005). The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. The red eyespot in Chlamydomonas is sensitive to light and hence determines movement. Genes & Dev, 19:176-187 [2]
- ^ Juliet Brodie & Jane Lewis (2007). Unravelling the algae: the past, present, and future of algal systematics. CRC Press. p. 140, [3].
- ^ Wehr, J.D., Sheath, R.G. & Kociolek, J.P. (eds., 2015). Freshwater Algae of North America: Ecology and Classification. Academic Press, USA, p. 275-276, [4].
- ^ Proschold, T.; Marin, B.; Schlösser, U. G.; Melkonian, M. (2001). "Molecular phylogeny and taxonomic revision of (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and Description of Oogamochlamys gen. nov. and Lobochlamys gen. nov". Protist. 152 (4): 265–300. doi:10.1078/1434-4610-00068. PMID 11822658.