| Clostridium perfringens | |
|---|---|
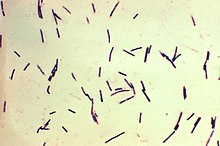
| |
| Photomicrograph of Gram-positive Clostridium perfringens bacilli | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Bacillota |
| Class: | Clostridia |
| Order: | Eubacteriales |
| Family: | Clostridiaceae |
| Genus: | Clostridium |
| Species: | C. perfringens
|
| Binomial name | |
| Clostridium perfringens Veillon & Zuber 1898
Hauduroy et al. 1937 | |
Clostridium perfringens (formerly known as C. welchii, or Bacillus welchii) is a Gram-positive, bacillus (rod-shaped), anaerobic, spore-forming pathogenic bacterium of the genus Clostridium.[1][2] C. perfringens is ever-present in nature and can be found as a normal component of decaying vegetation, marine sediment, the intestinal tract of humans and other vertebrates, insects, and soil. It has the shortest reported generation time of any organism at 6.3 minutes in thioglycolate medium.[3]
Clostridium perfringens is one of the most common causes of food poisoning in the United States, alongside norovirus, Salmonella, Campylobacter, and Staphylococcus aureus.[4] However, it can sometimes be ingested and cause no harm.[5]
Infections induced by C. perfringens are associated with tissue necrosis, bacteremia, emphysematous cholecystitis, and gas gangrene, which is also known as clostridial myonecrosis.[6] The specific name, perfringens, is derived from the Latin per (meaning "through") and frango ("burst"), referring to the disruption of tissue that occurs during gas gangrene.[7] Gas gangrene is caused by alpha toxin, or α-toxin, that embeds itself into the plasma membrane of cells and disrupts normal cellular function by altering membrane structure.[8] Research suggests that C. perfringens is capable of engaging in polymicrobial anaerobic infections.[9] It is commonly encountered in infections as a component of the normal flora. In this case, its role in disease is minor.[10]
C. perfringens toxins are a result of horizontal gene transfer of a neighboring cell's plasmids.[11] Shifts in genomic make-up are common for this species of bacterium and contribute to novel pathogenesis.[12] Major toxins are expressed differently in certain populations of C. perfringens; these populations are organized into strains based on their expressed toxins.[13] This especially impacts the food industry, as controlling this microbe is important for preventing foodborne illness.[12] Novel findings in C. perfringens hyper-motility, which was provisionally thought as non-motile, have been discovered as well.[14] Findings in metabolic processes reveal more information concerning C. perfringens pathogenic nature.[15]
- ^ Ryan, Kenneth J.; Ray, C. George (2004). Sherris Medical Microbiology : an Introduction to Infectious Diseases (4th ed.). New York: McGraw-Hill. p. 310. ISBN 978-0-8385-8529-0.
- ^ Kiu, R; Hall, L. J. (2018). "An update on the human and animal enteric pathogen Clostridium perfringens". Emerging Microbes & Infections. 7 (141): 141. doi:10.1038/s41426-018-0144-8. PMC 6079034. PMID 30082713.
- ^ "BioNumber Details Page". BioNumbers.
- ^ "Foodborne Illnesses and Germs". Centers for Disease Control and Prevention (CDC). 2018-02-16. Retrieved 18 February 2018.
- ^ Juckett, G; Bardwell, G; McClane, B; Brown, S (2008). "Microbiology of salt rising bread". The West Virginia Medical Journal. 104 (4): 26–7. PMID 18646681.
- ^ Hendrix, Nancy; Mackeen, A.; Weiner, Stuart (2011-01-24). "Clostridium perfringens Sepsis and Fetal Demise after Genetic Amniocentesis". American Journal of Perinatology Reports. 1 (1): 025–028. doi:10.1055/s-0030-1271221. ISSN 2157-6998. PMC 3653538. PMID 23705080.
- ^ Lexicon Orthopaedic Etymology. CRC Press. 1999. p. 128. ISBN 9789057025976.
- ^ Hendrix, Nancy; Mackeen, A.; Weiner, Stuart (2011-01-24). "Clostridium perfringens Sepsis and Fetal Demise after Genetic Amniocentesis". American Journal of Perinatology Reports. 1 (1): 025–028. doi:10.1055/s-0030-1271221. ISSN 2157-6998. PMC 3653538. PMID 23705080.
- ^ Takehara, Masaya; Takagishi, Teruhisa; Seike, Soshi; Ohtani, Kaori; Kobayashi, Keiko; Miyamoto, Kazuaki; Shimizu, Tohru; Nagahama, Masahiro (2016-06-16). "Clostridium perfringens α-Toxin Impairs Innate Immunity via Inhibition of Neutrophil Differentiation". Scientific Reports. 6 (1): 28192. Bibcode:2016NatSR...628192T. doi:10.1038/srep28192. ISSN 2045-2322. PMC 4910053. PMID 27306065.
- ^ Grenda, Tomasz; Jarosz, Aleksandra; Sapała, Magdalena; Grenda, Anna; Patyra, Ewelina; Kwiatek, Krzysztof (2023-05-26). "Clostridium perfringens—Opportunistic Foodborne Pathogen, Its Diversity and Epidemiological Significance". Pathogens. 12 (6): 768. doi:10.3390/pathogens12060768. ISSN 2076-0817. PMC 10304509. PMID 37375458.
- ^ Cite error: The named reference
Gulliver-2023was invoked but never defined (see the help page). - ^ a b Cite error: The named reference
Elnar-2021was invoked but never defined (see the help page). - ^ Revitt-Mills, Sarah A; Rood, Julian I; Adams, Vicki (2015). "Clostridium perfringens extracellular toxins and enzymes: 20 and counting". Microbiology Australia. 36 (3): 114. doi:10.1071/MA15039. ISSN 1324-4272.
- ^ Wambui, Joseph; Cernela, Nicole; Stevens, Marc J. A.; Stephan, Roger (2021-09-13). "Whole Genome Sequence-Based Identification of Clostridium estertheticum Complex Strains Supports the Need for Taxonomic Reclassification Within the Species Clostridium estertheticum". Frontiers in Microbiology. 12. doi:10.3389/fmicb.2021.727022. ISSN 1664-302X. PMC 8473909. PMID 34589074.
- ^ Cite error: The named reference
Ohtani-2016was invoked but never defined (see the help page).