A corynebacteriophage (or just corynephage) is a DNA-containing bacteriophage specific for bacteria of genus Corynebacterium as its host.[1] Corynebacterium diphtheriae virus[2] strain Corynebacterium diphtheriae phage[2] (aka Corynephage β[3] or just β-phage[4][5]) introduces toxigenicity into strains of Corynebacterium diphtheriae as it encodes diphtheria toxin,[6][7][8][5][4] it has subtypes beta c and beta vir.[9] According to proposed taxonomic classification, corynephages β and ω are unclassified members of the genus Lambdavirus, family Siphoviridae.[3][10]

Corynebacteriophage play a crucial role in the ecology and evolution of Corynebacterium species. They are viruses that specifically target and infect these bacteria, injecting their genetic material into the bacterial host and using the host's cellular machinery to replicate and produce more phages. The study of Corynephages is important not only for understanding the biology of these viruses but also for potential applications in biotechnology, such as using phages for bacterial control.
Corynebacteriophage, like other bacteriophages, exhibit a diverse range of structures and are classified based on various characteristics, including morphology, nucleic acid type, and genetic content.
Structure and Classification
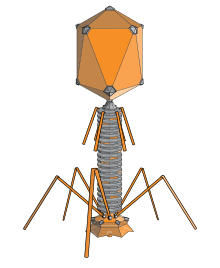
Corynephages are a diverse group of viruses that specifically infect bacteria within the Corynebacterium genus. Structurally, these phages exhibit a variety of forms, with most having a distinct head-and-tail morphology characteristic of the order Caudovirales. The capsid, typically icosahedral in shape, encloses the genetic material, which can be either double-stranded or single-stranded DNA, varying in size among different phage species. The tail structure, used for attachment to host cells, can be short or long, and its complexity varies across different phages. The specific morphology of a corynephage plays a key role in its classification and is a crucial factor in understanding its interaction with host bacteria.
| General overview of the structure and classification of Corynebacteriophages: | |
|---|---|
| Head (Capsid): | The head or capsid of a Corynebacteriophage contains the genetic material (DNA or RNA). It is often icosahedral in shape. |
| Tail: | Many Corynebacteriophages have a tail structure that helps in attaching to the bacterial host and injecting the genetic material. The tail can be short or long, depending on the specific phage. |
| Tail Fibers: | Some phages have tail fibers that aid in the recognition and attachment to specific receptors on the bacterial cell surface. |
| Base Plate: | The base plate is a structure at the base of the tail that may contain proteins involved in host cell recognition and the initiation of infection. |
Genetic Material:
DNA Phages: Most Corynebacteriophages have DNA as their genetic material. The DNA can be single-stranded or double-stranded. RNA Phages: Some phages, though less common, have RNA genomes.
Morphology:
Siphoviridae: Phages in this family typically have long, non-contractile tails and are characterized by their ability to undergo lysogeny (integration of their DNA into the host genome).
Myoviridae: These phages have long, contractile tails and are often associated with a lytic life cycle.
Podoviridae: Phages in this family have short tails.
Life Cycles and Host Interaction
Corynephages exhibit two primary life cycles: lytic and lysogenic. In the lytic cycle, the phage attaches to the bacterial cell, injects its DNA, and uses the cell's machinery to replicate its genome and produce new virions. This process eventually leads to the lysis of the host cell and the release of new phage particles. In contrast, the lysogenic cycle involves the integration of the phage's genetic material into the host's genome, forming a prophage. This integrated DNA is replicated along with the host's genome during cell division. The prophage may eventually enter the lytic cycle in response to specific triggers, such as stress or UV radiation. The choice between these life cycles is influenced by various factors, including the environmental conditions and the genetic makeup of both the phage and the host. Host Specificity Phages are often classified based on their host range and specificity, indicating the particular bacterial species or strain they can infect.
Applications in Research and Medicine
Corynephages are of significant interest in the fields of microbiology and biotechnology. Their specificity to Corynebacterium species makes them valuable tools in the study of these bacteria, including pathogenic strains like Corynebacterium diphtheriae. Phages are used to understand bacterial genetics, evolution, and mechanisms of pathogenicity. In the era of increasing antibiotic resistance, corynephages also offer potential in phage therapy, an alternative to traditional antibiotics. Phage therapy exploits the natural ability of phages to infect and lyse specific bacteria, providing a targeted approach to treat bacterial infections, especially those resistant to conventional treatments. Additionally, the genetic diversity and adaptability of corynephages make them a rich source for biotechnological applications, such as the development of novel antibacterial agents or the delivery of therapeutic genes.
Genome Size
The genomes of corynephages can vary widely in size. They typically range from about 15,000 to 100,000 base pairs, although this can vary depending on the specific phage.
Corynebacterium phage Corlili[11]
Phage stock CL31 obtained from Felix D'Herelle Reference Center for Bacterial Viruses. CL31 was originally isolated on C. glutamicum ATCC 15990 Centre de Recherche de Biochimie et de Génétique Cellulaire
CNRS, 31062 Toulouse, France before 1985.
The original CL31 phage stock makes plaques on MB001, albeit at a lower plaque formation efficiency than on ATCC 15990. Plaques obtained on MB001, when picked and propagated on the same strain, resulted in very high titer phage stocks.
- ^ NCBI: Corynephages (list)
- ^ a b NCBI: Corynebacterium diphtheriae virus/phage (species)
- ^ a b NCBI: Corynephage beta (species)
- ^ a b SIB: Modulation of host virulence by virus. Expasy: ViralZone. Accessed 18 Feb 2021
- ^ a b SIB: Viral exotoxin. Expasy: ViralZone. Accessed 18 Feb 2021
- ^ Julie K. Segman, ed. (2006). Stedman's Medical Dictionary (28th ed.). Baltimore, Maryland: Lippincott Williams & Wilkins. p. 449. ISBN 978-0-7817-3390-8.
- ^ TABLE 1. Bacterial virulence properties altered by bacteriophages from Wagner PL, Waldor MK (August 2002). "Bacteriophage control of bacterial virulence". Infection and Immunity. 70 (8): 3985–93. doi:10.1128/IAI.70.8.3985-3993.2002. PMC 128183. PMID 12117903.
- ^ L. P. Johnson, M. A. Tomai, P. M. Schlievert: Bacteriophage Involvement in Group A Streptococcal Pyrogenic Exotoxin A Production. In: Journal Of Bacteriology, Volume 166, No. 2, May 1986, pp 623-627
- ^ Costa, J. J.; Michel, J. L.; Rappuoli, R; Murphy, J. R. (1981). "Restriction map of corynebacteriophages beta c and beta vir and physical localization of the diphtheria tox operon". Journal of Bacteriology. 148 (1): 124–30. doi:10.1128/JB.148.1.124-130.1981. PMC 216174. PMID 6270058.
- ^ NCBI: Corynephage omega (species)
- ^ "The Actinobacteriophage Database | Phage Corlili". phagesdb.org. Retrieved 2023-12-17.