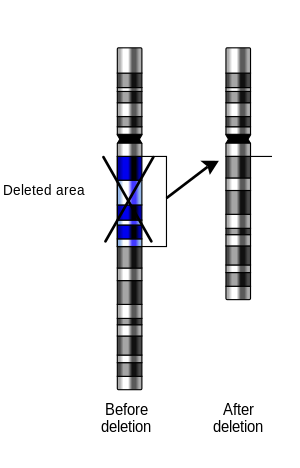
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome.[1] Some chromosomes have fragile spots where breaks occur, which result in the deletion of a part of the chromosome. The breaks can be induced by heat, viruses, radiation, or chemical reactions. When a chromosome breaks, if a part of it is deleted or lost, the missing piece of chromosome is referred to as a deletion or a deficiency.[2]
For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop.
The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand slippage, within the DNA polymerase active site.[3][4][5]
Deletions can be caused by errors in chromosomal crossover during meiosis, which causes several serious genetic diseases. Deletions that do not occur in multiples of three bases can cause a frameshift by changing the 3-nucleotide protein reading frame of the genetic sequence. Deletions are representative of eukaryotic organisms, including humans and not in prokaryotic organisms, such as bacteria.
- ^ Lewis, R. (2004). Human Genetics: Concepts and Applications (6th ed.). McGraw Hill. ISBN 978-0072951745.
- ^ Klug, William S. (2015). Concepts of genetics. Michael R. Cummings, Charlotte A. Spencer, Michael Angelo Palladino (Eleventh ed.). Boston. ISBN 978-0-321-94891-5. OCLC 880404074.
{{cite book}}: CS1 maint: location missing publisher (link) - ^ Banavali, Nilesh K. (2013). "Partial Base Flipping is Sufficient for Strand Slippage near DNA Duplex Termini". Journal of the American Chemical Society. 135 (22): 8274–8282. doi:10.1021/ja401573j. PMID 23692220.
- ^ Banavali, Nilesh K. (2013). "Analyzing the Relationship between Single Base Flipping and Strand Slippage near DNA Duplex Termini". The Journal of Physical Chemistry B. 117 (46): 14320–14328. doi:10.1021/jp408957c. PMID 24206351.
- ^ Manjari, Swati R.; Pata, Janice D.; Banavali, Nilesh K. (2014). "Cytosine Unstacking and Strand Slippage at an Insertion–Deletion Mutation Sequence in an Overhang-Containing DNA Duplex". Biochemistry. 53 (23): 3807–3816. doi:10.1021/bi500189g. PMC 4063443. PMID 24854722.