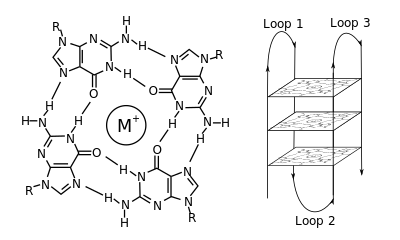
In molecular biology, G-quadruplex secondary structures (G4) are formed in nucleic acids by sequences that are rich in guanine.[2] They are helical in shape and contain guanine tetrads that can form from one,[3] two[4] or four strands.[5] The unimolecular forms often occur naturally near the ends of the chromosomes, better known as the telomeric regions, and in transcriptional regulatory regions of multiple genes, both in microbes[6][7] and across vertebrates [8][7] including oncogenes in humans.[9] Four guanine bases can associate through Hoogsteen hydrogen bonding to form a square planar structure called a guanine tetrad (G-tetrad or G-quartet), and two or more guanine tetrads (from G-tracts, continuous runs of guanine) can stack on top of each other to form a G-quadruplex.
The placement and bonding to form G-quadruplexes is not random and serve very unusual functional purposes. The quadruplex structure is further stabilized by the presence of a cation, especially potassium, which sits in a central channel between each pair of tetrads.[3] They can be formed of DNA, RNA, LNA, and PNA, and may be intramolecular, bimolecular, or tetramolecular.[10] Depending on the direction of the strands or parts of a strand that form the tetrads, structures may be described as parallel or antiparallel. G-quadruplex structures can be computationally predicted from DNA or RNA sequence motifs,[11][12] but their actual structures can be quite varied within and between the motifs, which can number over 100,000 per genome. Their activities in basic genetic processes are an active area of research in telomere, gene regulation, and functional genomics research.[13][14]
- ^ Capra JA, Paeschke K, Singh M, Zakian VA (July 2010). "G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae". PLOS Computational Biology. 6 (7): e1000861. Bibcode:2010PLSCB...6E0861C. doi:10.1371/journal.pcbi.1000861. PMC 2908698. PMID 20676380.
- ^ Routh ED, Creacy SD, Beerbower PE, Akman SA, Vaughn JP, Smaldino PJ (March 2017). "A G-quadruplex DNA-affinity Approach for Purification of Enzymaticacvly Active G4 Resolvase1". Journal of Visualized Experiments. 121 (121). doi:10.3791/55496. PMC 5409278. PMID 28362374.
- ^ a b Largy E, Mergny J, Gabelica V (2016). "Chapter 7. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability". In Astrid S, Helmut S, Roland KO (eds.). The Alkali Metal Ions: Their Role in Life (PDF). Metal Ions in Life Sciences. Vol. 16. Springer. pp. 203–258. doi:10.1007/978-3-319-21756-7_7. ISBN 978-3-319-21755-0. PMID 26860303.
- ^ Sundquist WI, Klug A (December 1989). "Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops". Nature. 342 (6251): 825–9. Bibcode:1989Natur.342..825S. doi:10.1038/342825a0. PMID 2601741. S2CID 4357161.
- ^ Sen D, Gilbert W (July 1988). "Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis". Nature. 334 (6180): 364–6. Bibcode:1988Natur.334..364S. doi:10.1038/334364a0. PMID 3393228. S2CID 4351855.
- ^ Rawal P, Kummarasetti VB, Ravindran R, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S (2006). "Genome-wide Prediction of G4 DNA as Regulatory Motifs: Role in Escherichia Coli Global Regulation". Genome Research. 16 (5): 644–655. doi:10.1101/gr.4508806. PMC 1457047. PMID 16651665.
- ^ a b Borman S (May 28, 2007). "Ascent of quadruplexes nucleic acid structures become promising drug targets". Chemical and Engineering News. 85 (22): 12–17. doi:10.1021/cen-v085n009.p012a.
- ^ Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S (2008). "Genome-wide Computational and Expression Analyses Reveal G-quadruplex DNA Motifs as Conserved Cis-Regulatory Elements in Human and Related Species". Journal of Medicinal Chemistry. 51 (18): 5641–5649. doi:10.1021/jm800448a. PMID 18767830.
- ^ Han H, Hurley LH (April 2000). "G-quadruplex DNA: a potential target for anti-cancer drug design". Trends in Pharmacological Sciences. 21 (4): 136–42. doi:10.1016/s0165-6147(00)01457-7. PMID 10740289.
- ^ Bochman ML, Paeschke K, Zakian VA (November 2012). "DNA secondary structures: stability and function of G-quadruplex structures". Nature Reviews. Genetics. 13 (11): 770–80. doi:10.1038/nrg3296. PMC 3725559. PMID 23032257.
- ^ Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S (2008). "QuadBase: Genome-Wide Database of G4 DNA--occurrence and Conservation in Human, Chimpanzee, Mouse and Rat Promoters and 146 Microbes". Nucleic Acids Research. 36 (Database): D381–D385. doi:10.1093/nar/gkm781. PMC 2238983. PMID 17962308.
- ^ Dhapola P, Chowdhury S (July 2016). "QuadBase2: Web Server for Multiplexed Guanine Quadruplex Mining and Visualization". Nucleic Acids Research. 44 (W1): W277–W283. doi:10.1093/nar/gkw425. PMC 4987949. PMID 27185890.
- ^ Rhodes D, Lipps HJ (October 2015). "G-quadruplexes and their regulatory roles in biology". Nucleic Acids Research. 43 (18): 8627–37. doi:10.1093/nar/gkv862. PMC 4605312. PMID 26350216.
- ^ Borman S (November 2009). "Promoter quadruplexes folded DNA structures in gene-activation sites may be useful cancer drug targets". Chemical and Engineering News. 87 (44): 28–30. doi:10.1021/cen-v087n044.p028.