| Glutathione synthetase | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Symbol | GSS | ||||||
| NCBI gene | 2937 | ||||||
| HGNC | 4624 | ||||||
| OMIM | 601002 | ||||||
| RefSeq | NM_000178 | ||||||
| UniProt | P48637 | ||||||
| Other data | |||||||
| EC number | 6.3.2.3 | ||||||
| Locus | Chr. 20 q11.2 | ||||||
| |||||||
| Eukaryotic glutathione synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Human glutathione synthetase | |||||||||
| Identifiers | |||||||||
| Symbol | GSH_synthase | ||||||||
| Pfam | PF03199 | ||||||||
| Pfam clan | CL0483 | ||||||||
| InterPro | IPR004887 | ||||||||
| SCOP2 | 2hgs / SCOPe / SUPFAM | ||||||||
| |||||||||
| glutathione synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
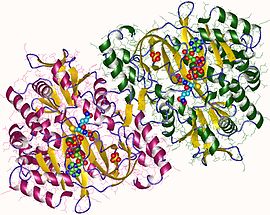 glutathione synthetase dimer, Human | |||||||||
| Identifiers | |||||||||
| EC no. | 6.3.2.3 | ||||||||
| CAS no. | 9023-62-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Eukaryotic glutathione synthase, ATP binding domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Human glutathione synthetase | |||||||||
| Identifiers | |||||||||
| Symbol | GSH_synth_ATP | ||||||||
| Pfam | PF03917 | ||||||||
| InterPro | IPR005615 | ||||||||
| SCOP2 | 1m0t / SCOPe / SUPFAM | ||||||||
| |||||||||
| Prokaryotic glutathione synthetase, N-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of escherichia coli glutathione synthetase at ph 7.5 | |||||||||
| Identifiers | |||||||||
| Symbol | GSH-S_N | ||||||||
| Pfam | PF02951 | ||||||||
| InterPro | IPR004215 | ||||||||
| SCOP2 | 1glv / SCOPe / SUPFAM | ||||||||
| |||||||||
| Prokaryotic glutathione synthetase, ATP-grasp domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of escherichia coli glutathione synthetase at ph 7.5 | |||||||||
| Identifiers | |||||||||
| Symbol | GSH-S_ATP | ||||||||
| Pfam | PF02955 | ||||||||
| Pfam clan | CL0179 | ||||||||
| InterPro | IPR004218 | ||||||||
| SCOP2 | 1glv / SCOPe / SUPFAM | ||||||||
| |||||||||
Glutathione synthetase (GSS) (EC 6.3.2.3) is the second enzyme in the glutathione (GSH) biosynthesis pathway. It catalyses the condensation of gamma-glutamylcysteine and glycine, to form glutathione.[2] Glutathione synthetase is also a potent antioxidant. It is found in many species including bacteria, yeast, mammals, and plants.[3]
In humans, defects in GSS are inherited in an autosomal recessive way and are the cause of severe metabolic acidosis, 5-oxoprolinuria, increased rate of haemolysis, and defective function of the central nervous system.[4] Deficiencies in GSS can cause a spectrum of deleterious symptoms in plants and human beings alike.[5]
In eukaryotes, this is a homodimeric enzyme. The substrate-binding domain has a three-layer alpha/beta/alpha structure.[6] This enzyme utilizes and stabilizes an acylphosphate intermediate to later perform a favorable nucleophilic attack of glycine.[citation needed]
- ^ Gogos A, Shapiro L (Dec 2002). "Large conformational changes in the catalytic cycle of glutathione synthase". Structure. 10 (12): 1669–76. doi:10.1016/S0969-2126(02)00906-1. PMID 12467574.
- ^ Njålsson R, Norgren S (2005). "Physiological and pathological aspects of GSH metabolism". Acta Paediatr. 94 (2): 132–7. doi:10.1111/j.1651-2227.2005.tb01878.x. PMID 15981742.
- ^ Li H, Xu H, Graham DE, White RH (Aug 2003). "Glutathione synthetase homologs encode alpha-L-glutamate ligases for methanogenic coenzyme F420 and tetrahydrosarcinapterin biosyntheses". Proceedings of the National Academy of Sciences of the United States of America. 100 (17): 9785–90. Bibcode:2003PNAS..100.9785L. doi:10.1073/pnas.1733391100. PMC 187843. PMID 12909715.
- ^ Cite error: The named reference
Njålsson_2005was invoked but never defined (see the help page). - ^ O'Neill M. "Glutathione Synthetase Deficiency". Online Mendelian Inheritance in Man. Archived from the original on 2016-02-10. Retrieved 2016-03-02.
- ^ Cite error: The named reference
Polekhina_1999was invoked but never defined (see the help page).