| Group I catalytic intron | |
|---|---|
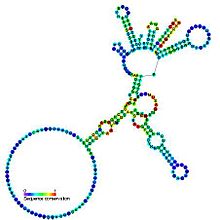 Predicted secondary structure and sequence conservation of Group I catalytic intron | |
| Identifiers | |
| Symbol | Intron_gpI |
| Rfam | RF00028 |
| Other data | |
| RNA type | Intron |
| Domain(s) | Eukaryota; Bacteria; Viruses |
| GO | GO:0000372 |
| SO | SO:0000587 |
| PDB structures | PDBe |
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms.[1][2][3] The core secondary structure consists of nine paired regions (P1-P9).[4] These fold to essentially two domains – the P4-P6 domain (formed from the stacking of P5, P4, P6 and P6a helices) and the P3-P9 domain (formed from the P8, P3, P7 and P9 helices).[2] The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.
- ^ Nielsen H, Johansen SD (2009). "Group I introns: Moving in new directions". RNA Biol. 6 (4): 375–83. doi:10.4161/rna.6.4.9334. PMID 19667762. Retrieved 2010-07-15.
- ^ a b Cate JH, Gooding AR, Podell E, et al. (September 1996). "Crystal structure of a group I ribozyme domain: principles of RNA packing". Science. 273 (5282): 1678–85. Bibcode:1996Sci...273.1678C. doi:10.1126/science.273.5282.1678. PMID 8781224. S2CID 38185676.
- ^ Cech TR (1990). "Self-splicing of group I introns". Annu. Rev. Biochem. 59: 543–68. doi:10.1146/annurev.bi.59.070190.002551. PMID 2197983.
- ^ Woodson SA (June 2005). "Structure and assembly of group I introns". Curr. Opin. Struct. Biol. 15 (3): 324–30. doi:10.1016/j.sbi.2005.05.007. PMID 15922592.