 | |
| Developer(s) | Sean Eddy, Travis Wheeler, HMMER development team |
|---|---|
| Stable release | 3.4[1]
/ 15 August 2023 |
| Repository | |
| Written in | C |
| Available in | English |
| Type | Bioinformatics tool |
| License | BSD-3 |
| Website | hmmer |
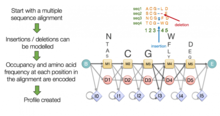
HMMER is a free and commonly used software package for sequence analysis written by Sean Eddy.[2] Its general usage is to identify homologous protein or nucleotide sequences, and to perform sequence alignments. It detects homology by comparing a profile-HMM (a Hidden Markov model constructed explicitly for a particular search) to either a single sequence or a database of sequences. Sequences that score significantly better to the profile-HMM compared to a null model are considered to be homologous to the sequences that were used to construct the profile-HMM. Profile-HMMs are constructed from a multiple sequence alignment in the HMMER package using the hmmbuild program. The profile-HMM implementation used in the HMMER software was based on the work of Krogh and colleagues.[3] HMMER is a console utility ported to every major operating system, including different versions of Linux, Windows, and macOS.
HMMER is the core utility that protein family databases such as Pfam and InterPro are based upon. Some other bioinformatics tools such as UGENE also use HMMER.
HMMER3 also makes extensive use of vector instructions to increase computational speed. This work is based upon an earlier publication showing a significant acceleration of the Smith-Waterman algorithm for aligning two sequences.[4]
- ^ "Release 3.4". 15 August 2023. Retrieved 18 September 2023.
- ^ Durbin, Richard; Sean R. Eddy; Anders Krogh; Graeme Mitchison (1998). Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press. ISBN 0-521-62971-3.
- ^ Krogh A, Brown M, Mian IS, Sjölander K, Haussler D (February 1994). "Hidden Markov models in computational biology. Applications to protein modeling". J. Mol. Biol. 235 (5): 1501–31. doi:10.1006/jmbi.1994.1104. PMID 8107089.
- ^ Farrar M (January 2007). "Striped Smith-Waterman speeds database searches six times over other SIMD implementations". Bioinformatics. 23 (2): 156–61. doi:10.1093/bioinformatics/btl582. PMID 17110365.