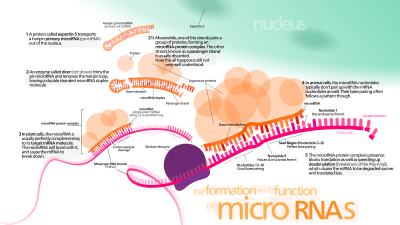

Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides.[1] Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression.[2][3] miRNAs base-pair to complementary sequences in messenger RNA (mRNA) molecules,[4] then silence said mRNA molecules by one or more of the following processes:[1][5]
- Cleaving the mRNA strand into two pieces.
- Destabilizing the mRNA by shortening its poly(A) tail.
- Reducing translation of the mRNA into proteins.
In cells of humans and other animals, miRNAs primarily act by destabilizing the mRNA.[6][7]
miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short hairpins, whereas siRNAs derive from longer regions of double-stranded RNA.[2] The human genome may encode over 1900 miRNAs,[8][9] However, only about 500 human miRNAs represent bona fide miRNAs in the manually curated miRNA gene database MirGeneDB.[10]
miRNAs are abundant in many mammalian cell types.[11][12] They appear to target about 60% of the genes of humans and other mammals.[13][14] Many miRNAs are evolutionarily conserved, which implies that they have important biological functions.[15][1] For example, 90 families of miRNAs have been conserved since at least the common ancestor of mammals and fish, and most of these conserved miRNAs have important functions, as shown by studies in which genes for one or more members of a family have been knocked out in mice.[1]
In 2024, American scientists Victor Ambros and Gary Ruvkun were awarded the Nobel Prize in Physiology or Medicine for their work on the discovery of miRNA and its role in post-transcriptional gene regulation.[16][17][18]
- ^ a b c d Bartel DP (March 2018). "Metazoan MicroRNAs". Cell. 173 (1): 20–51. doi:10.1016/j.cell.2018.03.006. PMC 6091663. PMID 29570994.
- ^ a b Bartel DP (January 2004). "MicroRNAs: genomics, biogenesis, mechanism, and function". Cell. 116 (2): 281–297. doi:10.1016/S0092-8674(04)00045-5. PMID 14744438.
- ^ Qureshi A, Thakur N, Monga I, Thakur A, Kumar M (January 2014). "VIRmiRNA: A comprehensive resource for experimentally validated viral miRNAs and their targets". Database. 2014: bau103. doi:10.1093/database/bau103. PMC 4224276. PMID 25380780.
- ^ Bartel DP (January 2009). "MicroRNAs: Target recognition and regulatory functions". Cell. 136 (2): 215–233. doi:10.1016/j.cell.2009.01.002. PMC 3794896. PMID 19167326.
- ^ Jonas S, Izaurralde E (July 2015). "Towards a molecular understanding of microRNA-mediated gene silencing". Nature Reviews. Genetics. 16 (7): 421–433. doi:10.1038/nrg3965. PMID 26077373. S2CID 24892348.
- ^ Jonas S, Izaurralde E (July 2015). "Towards a molecular understanding of microRNA-mediated gene silencing". Nature Reviews. Genetics. 16 (7): 421–433. doi:10.1038/nrg3965. PMID 26077373. S2CID 24892348.
- ^ Guo H, Ingolia NT, Weissman JS, Bartel DP (August 2010). "Mammalian microRNAs predominantly act to decrease target mRNA levels". Nature. 466 (7308): 835–840. Bibcode:2010Natur.466..835G. doi:10.1038/nature09267. hdl:1721.1/72447. PMC 2990499. PMID 20703300.
- ^ "Homo sapiens miRNAs". miRBase. Manchester, UK: Manchester University.
- ^ Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, et al. (April 2019). "An estimate of the total number of true human miRNAs". Nucleic Acids Research. 47 (7): 3353–3364. doi:10.1093/nar/gkz097. PMC 6468295. PMID 30820533.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ Fromm B, Domanska D, Høye E, Ovchinnikov V, Kang W, Aparicio-Puerta E, et al. (January 2020). "MirGeneDB 2.0: The metazoan microRNA complement". Nucleic Acids Research. 48 (D1): D132–D141. doi:10.1093/nar/gkz885. PMC 6943042. PMID 31598695.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, et al. (April 2003). "The microRNAs of Caenorhabditis elegans". Genes & Development. 17 (8): 991–1008. doi:10.1101/gad.1074403. PMC 196042. PMID 12672692.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (April 2002). "Identification of tissue-specific microRNAs from mouse". Current Biology. 12 (9): 735–739. Bibcode:2002CBio...12..735L. doi:10.1016/S0960-9822(02)00809-6. PMID 12007417.
- ^ Lewis BP, Burge CB, Bartel DP (January 2005). "Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets". Cell. 120 (1): 15–20. doi:10.1016/j.cell.2004.12.035. PMID 15652477.
- ^ Friedman RC, Farh KK, Burge CB, Bartel DP (January 2009). "Most mammalian mRNAs are conserved targets of microRNAs". Genome Research. 19 (1): 92–105. doi:10.1101/gr.082701.108. PMC 2612969. PMID 18955434.
- ^ Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, et al. (2015). "A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome". Annual Review of Genetics. 49 (1): 213–242. Bibcode:2015ARGen..49..213F. doi:10.1146/annurev-genet-120213-092023. PMC 4743252. PMID 26473382.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ "The Nobel Prize in Physiology or Medicine 2024".
- ^ "The Nobel Prize in Physiology or Medicine 2024". NobelPrize.org (Press release). Retrieved 7 October 2024.
- ^ Lewis T (7 October 2024). "Nobel Prize in Physiology or Medicine awarded for discovery of microRNA gene regulation". Scientific American. Retrieved 7 October 2024.