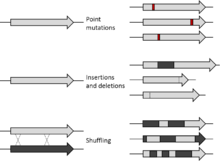
In molecular biology, mutagenesis is an important laboratory technique whereby DNA mutations are deliberately engineered to produce libraries of mutant genes, proteins, strains of bacteria, or other genetically modified organisms. The various constituents of a gene, as well as its regulatory elements and its gene products, may be mutated so that the functioning of a genetic locus, process, or product can be examined in detail. The mutation may produce mutant proteins with interesting properties or enhanced or novel functions that may be of commercial use. Mutant strains may also be produced that have practical application or allow the molecular basis of a particular cell function to be investigated.
Many methods of mutagenesis exist today. Initially, the kind of mutations artificially induced in the laboratory were entirely random using mechanisms such as UV irradiation. Random mutagenesis cannot target specific regions or sequences of the genome; however, with the development of site-directed mutagenesis, more specific changes can be made. Since 2013, development of the CRISPR/Cas9 technology, based on a prokaryotic viral defense system, has allowed for the editing or mutagenesis of a genome in vivo.[1] Site-directed mutagenesis has proved useful in situations that random mutagenesis is not. Other techniques of mutagenesis include combinatorial and insertional mutagenesis. Mutagenesis that is not random can be used to clone DNA,[2] investigate the effects of mutagens,[3] and engineer proteins.[4] It also has medical applications such as helping immunocompromised patients, research and treatment of diseases including HIV and cancers, and curing of diseases such as beta thalassemia.[5]
- ^ Hsu PD, Lander ES, Zhang F (June 2014). "Development and applications of CRISPR-Cas9 for genome engineering". Cell. 157 (6): 1262–78. doi:10.1016/j.cell.2014.05.010. PMC 4343198. PMID 24906146.
- ^ Motohashi K (June 2015). "A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis". BMC Biotechnology. 15: 47. doi:10.1186/s12896-015-0162-8. PMC 4453199. PMID 26037246.
- ^ Doering JA, Lee S, Kristiansen K, Evenseth L, Barron MG, Sylte I, LaLone CA (November 2018). "In Silico Site-Directed Mutagenesis Informs Species-Specific Predictions of Chemical Susceptibility Derived From the Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) Tool". Toxicological Sciences. 166 (1): 131–145. doi:10.1093/toxsci/kfy186. PMC 6390969. PMID 30060110.
- ^ Choi GC, Zhou P, Yuen CT, Chan BK, Xu F, Bao S, et al. (August 2019). "Combinatorial mutagenesis en masse optimizes the genome editing activities of SpCas9". Nature Methods. 16 (8): 722–730. doi:10.1038/s41592-019-0473-0. PMID 31308554. S2CID 196811756.
- ^ Bushman FD (February 2020). "Retroviral Insertional Mutagenesis in Humans: Evidence for Four Genetic Mechanisms Promoting Expansion of Cell Clones". Molecular Therapy. 28 (2): 352–356. doi:10.1016/j.ymthe.2019.12.009. PMC 7001082. PMID 31951833.