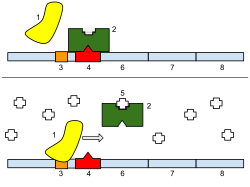
Bottom: The gene is turned on. Lactose is inhibiting the repressor, allowing the RNA polymerase to bind with the promoter and express the genes, which synthesize lactase. Eventually, the lactase will digest all of the lactose, until there is none to bind to the repressor. The repressor will then bind to the operator, stopping the manufacture of lactase.
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism.[1][2]
Promoters control gene expression in bacteria and eukaryotes.[3] RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters.
Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All DNAs have "Closely spaced promoters". Divergent, tandem, and convergent orientations are possible. Two closely spaced promoters will likely interfere. Regulatory elements can be several kilobases away from the transcriptional start site in gene promoters (enhancers).
In eukaryotes, the transcriptional complex can bend DNA, allowing regulatory sequences to be placed far from the transcription site. The distal promoter is upstream of the gene and may contain additional regulatory elements with a weaker influence. RNA polymerase II (RNAP II) bound to the transcription start site promoter can start mRNA synthesis. It also typically contains CpG islands, a TATA box, and TFIIB recognition elements.
Hypermethylation downregulates both genes, while demethylation upregulates them. Non-coding RNAs are linked to mRNA promoter regions. Subgenomic promoters range from 24 to 100 nucleotides (Beet necrotic yellow vein virus). Gene expression depends on promoter binding. Unwanted gene changes can increase a cell's cancer risk.
MicroRNA promoters often contain CpG islands. DNA methylation forms 5-methylcytosines at the 5' pyrimidine ring of CpG cytosine residues. Some cancer genes are silenced by mutation, but most are silenced by DNA methylation. Others are regulated promoters. Selection may favor less energetic transcriptional binding.
Variations in promoters or transcription factors cause some diseases. Misunderstandings can result from using a canonical sequence to describe a promoter.
- ^ Sharan R (4 January 2007). "Analysis of Biological Networks: Transcriptional Networks – Promoter Sequence Analysis" (PDF). Tel Aviv University. Retrieved 30 December 2012.
- ^ LaFleur TL, Hossain A, Salis HM (September 2022). "Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria". Nature Communications. 13 (1): 5159. Bibcode:2022NatCo..13.5159L. doi:10.1038/s41467-022-32829-5. PMC 9440211. PMID 36056029.
- ^ Vaishnav ED, de Boer CG, Molinet J, Yassour M, Fan L, Adiconis X, et al. (March 2022). "The evolution, evolvability and engineering of gene regulatory DNA". Nature. 603 (7901): 455–463. Bibcode:2022Natur.603..455V. doi:10.1038/s41586-022-04506-6. PMC 8934302. PMID 35264797.