| Squalene-hopene cyclase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
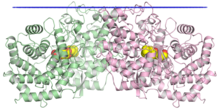 The crystallographic structure of the squalene-hopene cyclase dimer, with the membrane position indicated in blue, the two monomers in green and pink, and a substrate mimetic in the central cavity in yellow.[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 5.4.99.17 | ||||||||
| CAS no. | 76600-69-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Squalene-hopene cyclase (SHC) (EC 5.4.99.17) or hopan-22-ol hydro-lyase is an enzyme in the terpene cyclase/mutase family. It catalyzes the interconversion of squalene into a pentacyclic triterpenes, hopene and hopanol.[2][3][4][5][6] This enzyme catalyses the following chemical reactions.
- squalene hop-22(29)-ene
- squalene + H2O hopan-22-ol
SHC is important because its products, hopanoids, are very much like sterols in eukaryotes in that they condense lipid membranes and reduce permeability. In fact, SHC and sterol-producing enzymes (oxidosqualene cyclase) are evolutionarily related to each other.[7] Hopanoids are inferred to provide stability in the face of high temperatures and extreme acidity due to the rigid ring structure.[8] Indeed, up-regulation of SHC occurs in certain bacteria in the presence of hot or acidic environments.[9][10] SHC is found mostly in bacteria, but some eukaryotes, such as fungi and land plants, are also known to possess the enzyme.

- ^ Cite error: The named reference
wendtwas invoked but never defined (see the help page). - ^ Hoshino T, Sato T (February 2002). "Squalene-hopene cyclase: catalytic mechanism and substrate recognition". Chemical Communications (4): 291–301. doi:10.1039/b108995c. PMID 12120044.
- ^ Hoshino T, Nakano S, Kondo T, Sato T, Miyoshi A (May 2004). "Squalene-hopene cyclase: final deprotonation reaction, conformational analysis for the cyclization of (3R,S)-2,3-oxidosqualene and further evidence for the requirement of an isopropylidene moiety both for initiation of the polycyclization cascade and for the formation of the 5-membered E-ring". Organic & Biomolecular Chemistry. 2 (10): 1456–70. doi:10.1039/b401172d. PMID 15136801.
- ^ Sato T, Kouda M, Hoshino T (March 2004). "Site-directed mutagenesis experiments on the putative deprotonation site of squalene-hopene cyclase from Alicyclobacillus acidocaldarius". Bioscience, Biotechnology, and Biochemistry. 68 (3): 728–38. doi:10.1271/bbb.68.728. PMID 15056909.
- ^ Reinert DJ, Balliano G, Schulz GE (January 2004). "Conversion of squalene to the pentacarbocyclic hopene". Chemistry & Biology. 11 (1): 121–6. doi:10.1016/j.chembiol.2003.12.013. PMID 15113001.
- ^ Pearson A, Budin M, Brocks JJ (December 2003). "Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus". Proceedings of the National Academy of Sciences of the United States of America. 100 (26): 15352–7. Bibcode:2003PNAS..10015352P. doi:10.1073/pnas.2536559100. PMC 307571. PMID 14660793.
- ^ Santana-Molina, Carlos; Rivas-Marin, Elena; Rojas, Ana M; Devos, Damien P (2020-07-01). Ursula Battistuzzi, Fabia (ed.). "Origin and Evolution of Polycyclic Triterpene Synthesis". Molecular Biology and Evolution. 37 (7): 1925–1941. doi:10.1093/molbev/msaa054. ISSN 0737-4038. PMC 7306690. PMID 32125435.
- ^ Kannenberg, E.; Poralla, K. (1999). "Hopanoid biosynthesis and function in bacteria". Naturwissenschaften. 86 (4): 168–176. Bibcode:1999NW.....86..168K. doi:10.1007/s001140050592. S2CID 21596134.
- ^ Ourisson G, Rohmer M, Poralla K (1987). "Prokaryotic hopanoids and other polyterpenoid sterol surrogates". Annual Review of Microbiology. 41: 301–33. doi:10.1146/annurev.mi.41.100187.001505. PMID 3120639.
- ^ Sahm H, Rohmer M, Bringer-Meyer S, Sprenger GA, Welle R (1993). "Biochemistry and physiology of hopanoids in bacteria". Advances in Microbial Physiology. 35: 247–73. doi:10.1016/s0065-2911(08)60100-9. ISBN 9780120277353. PMID 8310881.
