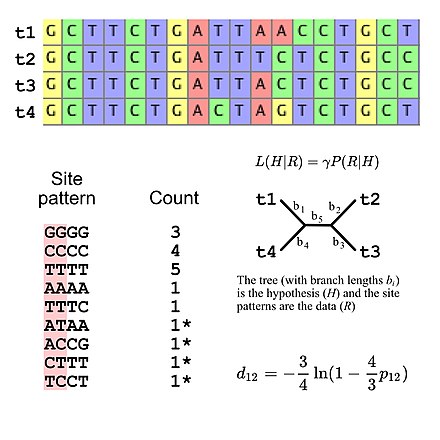
In biology, a substitution model, also called models of sequence evolution, are Markov models that describe changes over evolutionary time. These models describe evolutionary changes in macromolecules, such as DNA sequences or protein sequences, that can be represented as sequence of symbols (e.g., A, C, G, and T in the case of DNA or the 20 "standard" proteinogenic amino acids in the case of proteins). Substitution models are used to calculate the likelihood of phylogenetic trees using multiple sequence alignment data. Thus, substitution models are central to maximum likelihood estimation of phylogeny as well as Bayesian inference in phylogeny. Estimates of evolutionary distances (numbers of substitutions that have occurred since a pair of sequences diverged from a common ancestor) are typically calculated using substitution models (evolutionary distances are used input for distance methods such as neighbor joining). Substitution models are also central to phylogenetic invariants because they are necessary to predict site pattern frequencies given a tree topology. Substitution models are also necessary to simulate sequence data for a group of organisms related by a specific tree.