| Aldolase-type TIM barrel | |
|---|---|
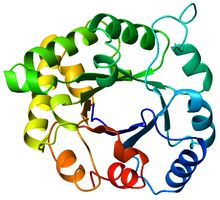 Top view of a triosephosphateisomerase (TIM) barrel (PDB: 8TIM), colored from blue (N-terminus) to red (C-terminus). | |
| Identifiers | |
| Symbol | Aldolase_TIM |
| Pfam clan | CL0036 |
| ECOD | 2002.1.1 |
| InterPro | IPR013785 |
| CATH | 8tim |
| SCOP2 | 8tim / SCOPe / SUPFAM |
The TIM barrel (triose-phosphate isomerase), also known as an alpha/beta barrel,[1]: 252 is a conserved protein fold consisting of eight alpha helices (α-helices) and eight parallel beta strands (β-strands) that alternate along the peptide backbone.[2] The structure is named after triose-phosphate isomerase, a conserved metabolic enzyme.[3] TIM barrels are ubiquitous, with approximately 10% of all enzymes adopting this fold.[4] Further, five of seven enzyme commission (EC) enzyme classes include TIM barrel proteins.[5][6] The TIM barrel fold is evolutionarily ancient, with many of its members possessing little similarity today,[7] instead falling within the twilight zone of sequence similarity.[8][9]
The inner beta barrel (β-barrel) is in many cases stabilized by intricate salt-bridge networks.[10] Loops at the C-terminal ends of the β-barrel are responsible for catalytic activity[11][12] while N-terminal end loops are important for the stability of the TIM-barrels. Structural inserts ranging from extended loops to independent protein domains may be inserted in place of these loops or at the N-terminus/C-terminals. TIM barrels appear to have evolved through gene duplication and domain fusion events of half-barrel proteins,[13] with a majority of TIM barrels originating from a common ancestor. This led many TIM barrels to possess internal symmetries.[14] Further gene duplication events of this ancestral TIM barrel led to diverging enzymes possessing the functional diversity observed today. TIM barrels have also been a longstanding target for protein designers. Successful TIM barrel designs include both domain fusions of existing proteins and de novo designs. Domain fusions experiments have resulted in many successful designs,[15][16][17][18][19][20][21] whereas de novo designs only yielded successes after 28 years of incremental development.[22]
- ^ Voet D, Voet JG (2011). "Chapter 8. Three-Dimensional Structures of Proteins". Biochemistry (4th ed.). John Wiley & Sons, Inc. ISBN 978-0470-91745-9.
- ^ Wierenga RK (March 2001). "The TIM-barrel fold: a versatile framework for efficient enzymes". FEBS Letters. 492 (3): 193–8. Bibcode:2001FEBSL.492..193W. doi:10.1016/s0014-5793(01)02236-0. PMID 11257493. S2CID 42044123.
- ^ Jansen R, Gerstein M (March 2000). "Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins". Nucleic Acids Research. 28 (6): 1481–8. doi:10.1093/nar/28.6.1481. PMC 111042. PMID 10684945.
- ^ Nagano N, Hutchinson EG, Thornton JM (October 1999). "Barrel structures in proteins: automatic identification and classification including a sequence analysis of TIM barrels". Protein Science. 8 (10): 2072–84. doi:10.1110/ps.8.10.2072. PMC 2144152. PMID 10548053.
- ^ Webb EC (1992). Enzyme nomenclature: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. Academic Press. ISBN 978-0-12-227164-9.
- ^ Nagano N, Orengo CA, Thornton JM (August 2002). "One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions". Journal of Molecular Biology. 321 (5): 741–65. doi:10.1016/s0022-2836(02)00649-6. PMID 12206759.
- ^ Livesay DR, La D (May 2005). "The evolutionary origins and catalytic importance of conserved electrostatic networks within TIM-barrel proteins". Protein Science. 14 (5): 1158–70. doi:10.1110/ps.041221105. PMC 2253277. PMID 15840824.
- ^ Chung SY, Subbiah S (October 1996). "A structural explanation for the twilight zone of protein sequence homology". Structure. 4 (10): 1123–7. doi:10.1016/s0969-2126(96)00119-0. PMID 8939745.
- ^ Vijayabaskar MS, Vishveshwara S (2012). "Insights into the fold organization of TIM barrel from interaction energy based structure networks". PLOS Computational Biology. 8 (5): e1002505. Bibcode:2012PLSCB...8E2505V. doi:10.1371/journal.pcbi.1002505. PMC 3355060. PMID 22615547.
- ^ Farber GK, Petsko GA (June 1990). "The evolution of alpha/beta barrel enzymes". Trends in Biochemical Sciences. 15 (6): 228–34. doi:10.1016/0968-0004(90)90035-A. PMID 2200166.
- ^ Reardon D, Farber GK (April 1995). "The structure and evolution of alpha/beta barrel proteins". FASEB Journal. 9 (7): 497–503. doi:10.1096/fasebj.9.7.7737457. PMID 7737457. S2CID 23208817.
- ^ Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M (September 2000). "Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion". Science. 289 (5484): 1546–50. Bibcode:2000Sci...289.1546L. doi:10.1126/science.289.5484.1546. PMID 10968789.
- ^ Söding J, Remmert M, Biegert A (July 2006). "HHrep: de novo protein repeat detection and the origin of TIM barrels". Nucleic Acids Research. 34 (Web Server issue): W137-42. doi:10.1093/nar/gkl130. PMC 1538828. PMID 16844977.
- ^ Seitz T, Bocola M, Claren J, Sterner R (September 2007). "Stabilisation of a (βα)8-barrel protein designed from identical half barrels". Journal of Molecular Biology. 372 (1): 114–29. doi:10.1016/j.jmb.2007.06.036. PMID 17631894.
- ^ Höcker B, Lochner A, Seitz T, Claren J, Sterner R (February 2009). "High-resolution crystal structure of an artificial (βα)(8)-barrel protein designed from identical half-barrels". Biochemistry. 48 (6): 1145–7. doi:10.1021/bi802125b. PMID 19166324.
- ^ Höcker B, Claren J, Sterner R, Makar AB, McMartin KE, Palese M, Tephly TR (June 1975). "Formate assay in body fluids: application in methanol poisoning". Biochemical Medicine. 13 (2): 117–26. doi:10.1016/0006-2944(75)90147-7. PMC 534502. PMID 15539462.
- ^ Claren J, Malisi C, Höcker B, Sterner R (March 2009). "Establishing wild-type levels of catalytic activity on natural and artificial (beta alpha)8-barrel protein scaffolds". Proceedings of the National Academy of Sciences of the United States of America. 106 (10): 3704–9. Bibcode:2009PNAS..106.3704C. doi:10.1073/pnas.0810342106. PMC 2656144. PMID 19237570.
- ^ Bharat TA, Eisenbeis S, Zeth K, Höcker B (July 2008). "A beta alpha-barrel built by the combination of fragments from different folds". Proceedings of the National Academy of Sciences of the United States of America. 105 (29): 9942–7. Bibcode:2008PNAS..105.9942B. doi:10.1073/pnas.0802202105. PMC 2481348. PMID 18632584.
- ^ Eisenbeis S, Proffitt W, Coles M, Truffault V, Shanmugaratnam S, Meiler J, Höcker B (March 2012). "Potential of fragment recombination for rational design of proteins". Journal of the American Chemical Society. 134 (9): 4019–22. doi:10.1021/ja211657k. PMID 22329686.
- ^ Fortenberry C, Bowman EA, Proffitt W, Dorr B, Combs S, Harp J, et al. (November 2011). "Exploring symmetry as an avenue to the computational design of large protein domains". Journal of the American Chemical Society. 133 (45): 18026–9. doi:10.1021/ja210593m. PMC 3781211. PMID 21978247.
- ^ Huang PS, Feldmeier K, Parmeggiani F, Velasco DA, Höcker B, Baker D (January 2016). "De novo design of a four-fold symmetric TIM-barrel protein with atomic-level accuracy". Nature Chemical Biology. 12 (1): 29–34. doi:10.1038/nchembio.1966. PMC 4684731. PMID 26595462.